Bessie Kebaara, PhD - PI

Bessie Kebaara, PhD
Associate Professor of Biology, Baylor University
Education:
BS: Microbiology, Louisiana State University, Baton Rouge, LA
PhD: Biological Sciences, University of Nebraska-Lincoln, NE
Courses Taught:
BIO 5100 RNA Biology
Research Interests
Regulation of gene expression in yeast
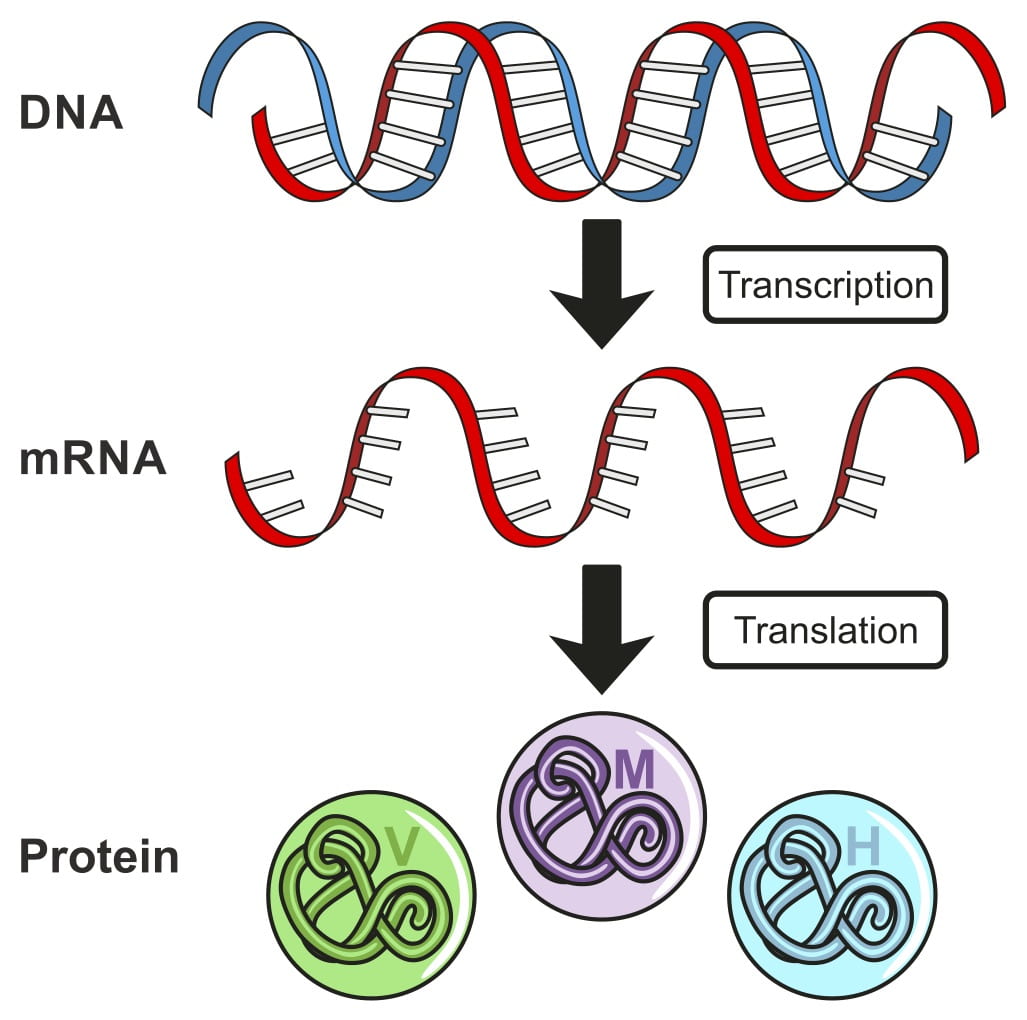
My research interests include the regulation of gene expression, specifically RNA biology. I am interested in understanding the recognition and targeting of natural mRNAs by the nonsense-mediated mRNA decay. The nonsense-mediated mRNA decay pathway (NMD) is an evolutionarily conserved mRNA surveillance pathway that contributes to the recognition and rapid degradation of mRNA with premature stop codons, thus preventing the accumulation of potentially harmful truncated proteins. This pathway also recognizes and degrades some natural physiological mRNAs. Natural mRNAs degraded by the nonsense mediated mRNA decay pathway have been identified in eukaryotes ranging from yeast (Saccharomyces cerevisiae.), to insects (Drosophila melanogaster), plants (Arabidopsis) and humans. My goal is to understand the recognition and targeting of natural mRNAs for NMD-mediated degradation, and understand the physiological consequences that are brought about by the degradation of specific mRNAs in the model organism S. cerevisiae.
Publications
Peccarelli, M., Scott, T.D., Kebaara, B.W., 2019. Nonsense-mediated mRNA decay of the ferric and cupric reductase mRNAs FRE1 and FRE2 in Saccharomyces cerevisiae. FEBS Letters 593, 3228–3238. https://doi.org/10.1002/1873-3468.13545
Murtha, K., Hwang, M., Peccarelli, M.C., Scott, T.D., Kebaara, B.W., 2019. The nonsense-mediated mRNA decay (NMD) pathway differentially regulates COX17, COX19 and COX23 mRNAs. Curr Genet 65, 507–521. https://doi.org/10.1007/s00294-018-0892-y
Peccarelli, M., Scott, T.D., Steele, M., Kebaara, B.W., 2016. mRNAs involved in copper homeostasis are regulated by the nonsense-mediated mRNA decay pathway depending on environmental conditions. Fungal Genetics and Biology 86, 81–90. https://doi.org/10.1016/j.fgb.2015.12.011
Obenoskey, J., Lane, D.R., Atkin, A.L., Kebaara, B.W., 2014. Immunity of the Saccharomyces cerevisiae SSY5 mRNA to nonsense-mediated mRNA decay. Front. Mol. Biosci. 1. https://doi.org/10.3389/fmolb.2014.00025
Peccarelli, M., Kebaara, B.W., 2014a. Measurement of mRNA Decay Rates in Saccharomyces cerevisiae Using rpb1-1 Strains. JoVE (Journal of Visualized Experiments) e52240. https://doi.org/10.3791/52240
Peccarelli, M., Kebaara, B.W., 2014b. Regulation of Natural mRNAs by the Nonsense-Mediated mRNA Decay Pathway. Eukaryotic Cell 13, 1126–1135. https://doi.org/10.1128/EC.00090-14
Peccarelli, M., Scott, T.D., Wong, H., Wang, X., Kebaara, B.W., 2014. Regulation of CTR2 mRNA by the nonsense-mediated mRNA decay pathway. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms 1839, 1283–1294. https://doi.org/10.1016/j.bbagrm.2014.09.011
Wang, X., Okonkwo, O., Kebaara, B.W., 2013. Physiological basis of copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Yeast 30, 179–190. https://doi.org/10.1002/yea.2950
Kebaara, B.W., Baker, K.E., Patefield, K.D., Atkin, A.L., 2012. Analysis of Nonsense-Mediated mRNA Decay in Saccharomyces cerevisiae. Current Protocols in Cell Biology 54, 27.3.1-27.3.39. https://doi.org/10.1002/0471143030.cb2703s54
Deliz-Aguirre, R., Atkin, A.L., Kebaara, B.W., 2011. Copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Curr Genet 57, 421–430. https://doi.org/10.1007/s00294-011-0356-0
Kebaara, B.W., Atkin, A.L., 2009. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 37, 2771–2778. https://doi.org/10.1093/nar/gkp146
Ghosh, S., Kebaara, B.W., Atkin, A.L., Nickerson, K.W., 2008. Regulation of Aromatic Alcohol Production in Candida albicans. Appl. Environ. Microbiol. 74, 7211–7218. https://doi.org/10.1128/AEM.01614-08
Kebaara, B.W., Langford, M.L., Navarathna, D.H.M.L.P., Dumitru, R., Nickerson, K.W., Atkin, A.L., 2008. Candida albicans Tup1 Is Involved in Farnesol-Mediated Inhibition of Filamentous-Growth Induction. Eukaryotic Cell 7, 980–987. https://doi.org/10.1128/EC.00357-07
Nazarenus, T., Cedarberg, R., Bell, R., Cheatle, J., Forch, A., Haifley, A., Hou, A., Wanja Kebaara, B., Shields, C., Stoysich, K., Taylor, R., Atkin, A.L., 2005. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116p. Gene 345, 199–212. https://doi.org/10.1016/j.gene.2004.10.005
Hornby, J.M., Kebaara, B.W., Nickerson, K.W., 2003. Farnesol Biosynthesis in Candida albicans: Cellular Response to Sterol Inhibition by Zaragozic Acid B. Antimicrobial Agents and Chemotherapy 47, 2366–2369. https://doi.org/10.1128/AAC.47.7.2366-2369.2003
Kebaara, Bessie, Nazarenus, T., Taylor, R., Atkin, A.L., 2003. Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr Genet 43, 171–177. https://doi.org/10.1007/s00294-003-0386-3
Kebaara, B., Nazarenus, T., Taylor, R., Forch, A., Atkin, A.L., 2003a. The Upf‐dependent decay of wild‐type PPR1 mRNA depends on its 5′‐UTR and first 92 ORF nucleotides. Nucleic Acids Res 31, 3157–3165. https://doi.org/10.1093/nar/gkg430
Kebaara, B., Nazarenus, T., Taylor, R., Forch, A., Atkin, A.L., 2003b. The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5′-UTR and first 92 ORF nucleotides. Nucleic Acids Res 31, 3157–3165.
Wang, X., Okonkwo, O., Kebaara, B.W., 2013. Physiological basis of copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Yeast 30, 179–190. https://doi.org/10.1002/yea.2950