Search Pubmed for publications from the Trakselis Laboratory. MyNCBI
From Trakselis Lab:
63) Behrmann, M.S., Perera, H.M., Welikala, M.U., Matthews, J.E, and Trakselis, M.A.* (2023) Dysregulated DnaB unwinding induces replisome decoupling and daughter strand gaps that are countered by RecA polymerization, under review.
62) Edwards, A.N., Blue, A.J., Conforti, J.M., Cordes, M.S., Trakselis, M.A., Gallagher, E.S.* (2023) Gas-phase stability and thermodynamics of ligand-bound, binary complexes of chloramphenicol acetyltransferase reveal negative cooperativity, Analy. Bioanaly. Chem., in press.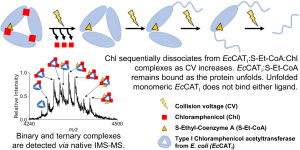 61) McKinzey, D., Li, C., Gao, Y., and and Trakselis, M.A.* (2023) Activity, Substrate Preference, and Structure of the HsMCM8/9 Helicase, NAR, Jun 13:gkad508. doi: 10.1093/nar/gkad508. PMID: 37309874.
61) McKinzey, D., Li, C., Gao, Y., and and Trakselis, M.A.* (2023) Activity, Substrate Preference, and Structure of the HsMCM8/9 Helicase, NAR, Jun 13:gkad508. doi: 10.1093/nar/gkad508. PMID: 37309874. 60) Griffin, W.C., McKinzey, D., Klinzing, K.A., Baratam, R., and Eliyapura, A., and Trakselis, M.A.* (2022) A multi-functional role for the MCM8/9 helicase complex in maintaining fork integrity during replication stress, Nat. Commun. 13, 5090. doi.org/10.1038/s41467-022-32583-8
60) Griffin, W.C., McKinzey, D., Klinzing, K.A., Baratam, R., and Eliyapura, A., and Trakselis, M.A.* (2022) A multi-functional role for the MCM8/9 helicase complex in maintaining fork integrity during replication stress, Nat. Commun. 13, 5090. doi.org/10.1038/s41467-022-32583-8

61) Trakselis, M.A. (ed) (2022) Helicase Enzymes Part B Methods in Enzymology. Vol 673
60) Trakselis, M.A. (ed) (2022) Helicase Enzymes Part A, Methods in Enzymology. Vol 672
 59) Behrmann, M.S. and Trakselis, M.A.* (2022) In vivo fluorescent TUNEL detection of single stranded DNA gaps and breaks induced by dnaB helicase mutants in Escherichia coli. Methods in Enzymology. Vol 672, 125-142.
59) Behrmann, M.S. and Trakselis, M.A.* (2022) In vivo fluorescent TUNEL detection of single stranded DNA gaps and breaks induced by dnaB helicase mutants in Escherichia coli. Methods in Enzymology. Vol 672, 125-142. 58) Kaszubowski, J.D. and Trakselis, M.A.* (2022) Beyond the lesion: Back to high fidelity DNA synthesis. Front. Mol. Biosci., Vol 8, 811540. link.
58) Kaszubowski, J.D. and Trakselis, M.A.* (2022) Beyond the lesion: Back to high fidelity DNA synthesis. Front. Mol. Biosci., Vol 8, 811540. link.
57) Behrmann, M.S., Perera, H.M., Hoang, J.M., Venkat, T.A., Visser, B.J., Bates, D. and
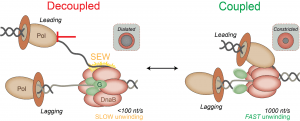
56) Perera, H.M. and

55)  54) A Unifying Framework for Understanding Biological Structures and Functions Across Levels of Biological Organization. Integr. Comp. Biol. icab167. link
54) A Unifying Framework for Understanding Biological Structures and Functions Across Levels of Biological Organization. Integr. Comp. Biol. icab167. link
 53) McKinzey, D.R., Gomathinayagam, S., Griffin, W.C., Klinzing, K.N., Jeffries, E.P., Rajkovic, A. and Trakselis, M.A. (2021) Motifs of the C-terminal domain of MCM9 direct localization to sites of mitomycin-C damage for RAD51 recruitment. JBC, 296, 100355. link bioRxiv
53) McKinzey, D.R., Gomathinayagam, S., Griffin, W.C., Klinzing, K.N., Jeffries, E.P., Rajkovic, A. and Trakselis, M.A. (2021) Motifs of the C-terminal domain of MCM9 direct localization to sites of mitomycin-C damage for RAD51 recruitment. JBC, 296, 100355. link bioRxiv

52) Cranford, M.T., Kaszubowski, J.D., and Trakselis, M.A. (2020) A hand-off of DNA between archaeal polymerases allows high-fidelity replication to resume at a discrete intermediate three bases past 8-oxoguanine. NAR, 48(19) 10986-97. link

51) Perera, H.M. and Trakselis, M.A. (2020) Site-specific DNA mapping using azidophenacyl bromide (APB) to map the binding orientation of proteins. Bio-protocol, 10(12): e3649. link
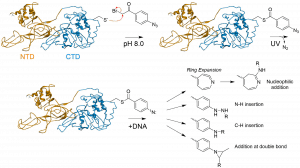
50) Perera, H.M. and Trakselis, M.A. (2019) Amidst multiple binding orientations on fork DNA, Saccharolobus MCM helicase proceeds N-first for unwinding. Elife, 8. pii: e46096. link

49) Perera, H.M., Behrmann, M.S., Hoang, J.M., Griffin, W.C., Trakselis, M.A. (2019) Contacts and context that regulate DNA helicase unwinding and replisome progression. The Enzymes, 45:183-223. link

48) Brosh, R.M. and Trakselis, M.A. (2019) Fine-tuning of the replisome: Mcm10 regulates fork progression and regression. Cell Cycle, 18(10):1047-1055. link
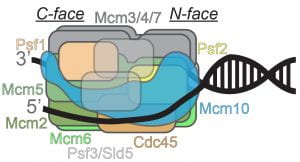
47) Griffin, W.C. and Trakselis, M.A. (2019) The MCM8/9 complex: A recent recruit to the roster of helicases involved in genome maintenance. DNA Repair, 76:1-10. link
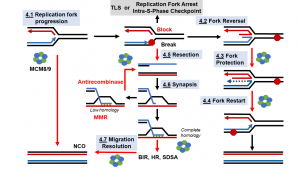
46) Graham, B.W., Perera, H., Bougoulias, M.E., Dodge, K.L., Thaxton, C.T., Olaso, D., Young, Tao, Y., N.L., Marshall, A.G., and Trakselis, M.A. (2018) Control of hexamerization, assembly, and excluded strand specificity for the Sulfolobus solfataricus MCM helicase., Biochemistry, 57, 5672-5682. link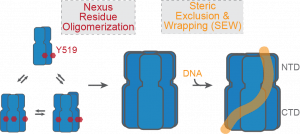
45) van Dongen, S. F. M., Clerx, J., van den Boomen, O. I., Pervaiz, M., Trakselis, M. A.
Ritschel, T., Schoonen, L., Schoenmakers, D. C. and Nolte, R. J. M. (2018) Synthetic polymers as substrates for a DNA-sliding clamp protein, Biopolymers, e23119 link

44) Carney, S.M., Gomathinayagam, S., Leuba, S.H., and Trakselis, M.A. (2017) Bacterial DnaB Helicase Interacts with the Excluded Strand to Regulate Unwinding, J. Biol. Chem., 292(46):19001-19012. doi: 10.1074/jbc.M117.814178.
43) Trakselis, M.A., Cranford, M.T., Chu, A.M. (2017) Coordination and Substitution of DNA Polymerases in Response to Genomic Obstacles, ACS Chem. Res. Tox., 30(11):1956-1971. doi: 10.1021/acs.chemrestox.7b00190.

42) Cranford, M.T., Chu, A.M., Baguley, J., Bauer, R.J., and Trakselis, M.A. (2017) Characterization of a Coupled DNA Replication and Translesion Synthesis Polymerase Supraholoenzyme, Nuc. Acids Res., 45, 8329-40. link
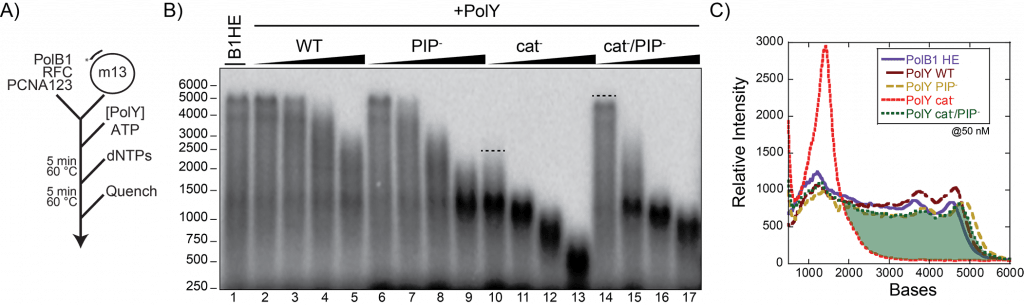
41) Trakselis, M.A., Siedman, M.M., Brosh, R.M. Jr. (2017) Mechanistic Insights Into How CMG Helicase Facilitates Replication Past DNA Roadblocks. DNA Repair, 108, 79-91. link
40) Khan, I., Crouch, J.D., Bharti, S.K., Sommers, J.A., Carney, S.M., Yakubovskaya, E., Garcia-Diaz, M., Trakselis, M.A., Brosh, R.M. Jr. (2016) Biochemical characterization of the human mitochondrial replicative Twinkle helicase: Substrate specificity, DNA branch-migration, and ability to overcome blockades to DNA unwinding. J. Biol. Chem., 291, 14324-39. link ![]()

39) Carney, S.M. and Trakselis, M.A. (2016) The Excluded DNA Strand is SEW Important for Hexameric Helicase Unwinding, Methods, 108, 79-91. doi:10.1016/j.ymeth.2016.04.008 in press. link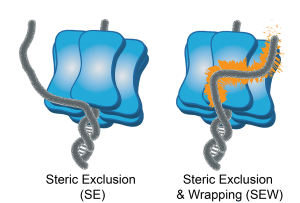 38) Graham, B.W., Tao, Y., Dodge, K.L., Zhang, Q., Thaxton, C.T., Olaso, D., Young, N.L., Marshal, A.G., and Trakselis, M.A. (2016) DNA Interactions probed by H/D exchange FT-ICR mass spectrometry confirm external surface binding sites on the MCM helicase. J. Biol. Chem., 291, 12467-80. link
38) Graham, B.W., Tao, Y., Dodge, K.L., Zhang, Q., Thaxton, C.T., Olaso, D., Young, N.L., Marshal, A.G., and Trakselis, M.A. (2016) DNA Interactions probed by H/D exchange FT-ICR mass spectrometry confirm external surface binding sites on the MCM helicase. J. Biol. Chem., 291, 12467-80. link
 37) Trakselis, M.A., (2016) Structural mechanisms of hexameric helicase loading, assembly, and unwinding. F1000Research, 5(F1000 Faculty Rev):111 (doi: 10.12688/f1000research.7509.1). link
37) Trakselis, M.A., (2016) Structural mechanisms of hexameric helicase loading, assembly, and unwinding. F1000Research, 5(F1000 Faculty Rev):111 (doi: 10.12688/f1000research.7509.1). link
 36) Barnes, C.O.,Calero, M.,Malik, I.,Graham, B.W., Spahr, H.,Lin, G.,Cohen, A.E., Brown, I.S.,Zhang, Q., Pullara, F., Trakselis, M.A., Kaplan, C.D., Calero, G. (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol. Cell, 59 (2), 258-69. link
36) Barnes, C.O.,Calero, M.,Malik, I.,Graham, B.W., Spahr, H.,Lin, G.,Cohen, A.E., Brown, I.S.,Zhang, Q., Pullara, F., Trakselis, M.A., Kaplan, C.D., Calero, G. (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol. Cell, 59 (2), 258-69. link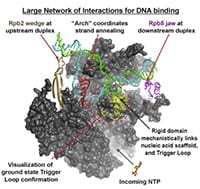 35) AlAsiri, S., Basit, S., Wood-Trageser, M.A., Yatsenko, S.A., Jeffries, E.P., Surti, U., Ketterer, D.M., Afzal, S., Ramzan, R., Faiyaz-Ul Haque, M., Jiang, H, Trakselis, M.A., Rajkovic, A. (2015) Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J. Clin. Invest., 125 (1), 754-62. link
35) AlAsiri, S., Basit, S., Wood-Trageser, M.A., Yatsenko, S.A., Jeffries, E.P., Surti, U., Ketterer, D.M., Afzal, S., Ramzan, R., Faiyaz-Ul Haque, M., Jiang, H, Trakselis, M.A., Rajkovic, A. (2015) Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J. Clin. Invest., 125 (1), 754-62. link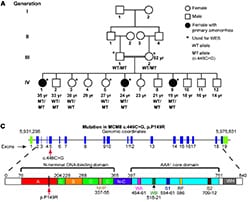 34) Wood-Trageser, M.A., Gurbuz, F., Yatsenko, S.A., Jeffries, E.P., Kotan, L.D., Surti, U., Ketterer, D.M., Matic, J., Chipkin, J., Jiang, H, Trakselis, M.A., Topaloglu, A.K., Rajkovic, A. (2014) Exome sequencing identifies MCM9 mutations in ovarian failure, short stature and chromosomal instability. Am. J. Hum. Genetics, 95 (6), 754-762. link
34) Wood-Trageser, M.A., Gurbuz, F., Yatsenko, S.A., Jeffries, E.P., Kotan, L.D., Surti, U., Ketterer, D.M., Matic, J., Chipkin, J., Jiang, H, Trakselis, M.A., Topaloglu, A.K., Rajkovic, A. (2014) Exome sequencing identifies MCM9 mutations in ovarian failure, short stature and chromosomal instability. Am. J. Hum. Genetics, 95 (6), 754-762. link 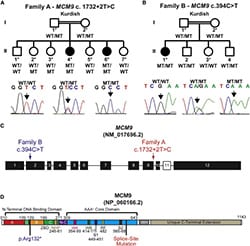 33) Nucleic Acid Polymerases, Murakami, K. and Trakselis, M.A. (Eds.), Nucleic Acids and Molecular Biology Vol. 30, Berlin, Germany, Springer. 2014. link
33) Nucleic Acid Polymerases, Murakami, K. and Trakselis, M.A. (Eds.), Nucleic Acids and Molecular Biology Vol. 30, Berlin, Germany, Springer. 2014. link
 32) Trakselis, M.A. and Murakami, K. Introduction to Nucleic Acid Polymerases: Families, Themes, and Mechanisms, (Chapter 1), Nucleic Acid Polymerases, Murakami, K. and Trakselis, M.A. (Eds.), Nucleic Acids and Molecular Biology Vol. 30, Berlin, Germany, Springer. 2014. link
32) Trakselis, M.A. and Murakami, K. Introduction to Nucleic Acid Polymerases: Families, Themes, and Mechanisms, (Chapter 1), Nucleic Acid Polymerases, Murakami, K. and Trakselis, M.A. (Eds.), Nucleic Acids and Molecular Biology Vol. 30, Berlin, Germany, Springer. 2014. link
31) Trakselis, M.A. and Bauer, R.J. Archaeal DNA Polymerases: Enzymatic Abilities, Coordination, and Unique Properties, (Chapter 6), Nucleic Acid Polymerases, Murakami, K. and Trakselis, M.A. (Eds.), Nucleic Acids and Molecular Biology Vol. 30, Berlin, Germany, Springer. 2014. link
30) Bauer, R.J., Wolff, I.D., Zuo, X., Lin, H-K., Trakselis, M.A. (2013) Assembly and Distributive Action of an Archaeal DNA Polymerase Holoenzyme. Special Issue of J. Mol. Biol. entitled “DNA replication & Genomic instability – Replicon Theory”, 425 (230), 4820-36. link
29) Mohan S., Das, D., Bauer R.J., Heroux A., Zalewski, J.K., Heber, S., Dosunmu-Oqunbi, A.M., Trakselis, M.A., Hildebrand J.D., and VanDemark A.P (2013) Structure of a Highly Conserved Domain of Rock1 Required for Shroom-mediated Regulation of Cell Morphology. PLOS One, 8(12), e81075. link
28) van Dongen, S.F.M., Clerx, J., Nørgaard, K., Bloemberg. T.G., Cornelissen, J.J.L.M., Trakselis, M.A., Nelson, S.W., Benkovic, S.J., Rowan, A.E., Nolte, R.J.M. A Clamp-Shaped Bio-Hybrid Catalyst for DNA Oxidation. Nature Chem., 5(11), 946-51. link
 Commentary by: Prins, L.J. & Scrimin, S. Processive Catalysis: Tread and Cut, News and Views, Nature Chem., 5, 899-900.
Commentary by: Prins, L.J. & Scrimin, S. Processive Catalysis: Tread and Cut, News and Views, Nature Chem., 5, 899-900.
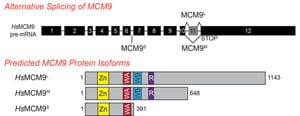
27) Jeffries, E.P., Denq, W.I., Bartko, J.C., and Trakselis, M.A. (2013) Identification, Quantification, and Evolutionary Analysis of a Novel Isoform of MCM9, Gene, 519, 41-49. link
26) Bauer, R.J., Graham, B.W., and Trakselis, M.A. (2013) Novel Interaction of the Bacterial-like DnaG Primase with the MCM Helicase in Archaea, J. Mol. Biol., 8(26), 1259-1273. link
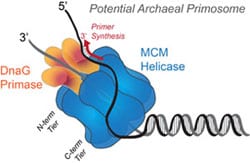
25) Trakselis, M.A. and Graham, B.W. (2012) Biochemisty: Molecular Hurdles Cleared with Ease, Nature, 492, 195-197. link
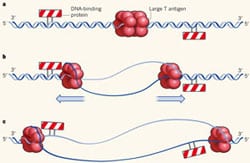
24) Lin, H-K., Chase, S.F., Laue, T.M., Jen-Jacobsen, L., and Trakselis, M.A. (2012) Differential Temperature Dependent Multimeric Assemblies of Replication and Repair Polymerases on DNA Increase Processivity. Biochemistry,51, 7367-7382. link
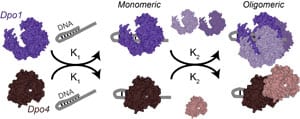 23) Mohan, S., Rizaldy, R., Das D., Bauer R.J., Heroux A., Trakselis, M.A., Hildebrand J.D., Vandemark, A.P. (2012) Structure of the shroom domain 2 reveals a three-segmented coiled-coil required for dimerization, rock binding, and apical constriction, Mol. Biol. Cell, 23, 2131-2142. link
23) Mohan, S., Rizaldy, R., Das D., Bauer R.J., Heroux A., Trakselis, M.A., Hildebrand J.D., Vandemark, A.P. (2012) Structure of the shroom domain 2 reveals a three-segmented coiled-coil required for dimerization, rock binding, and apical constriction, Mol. Biol. Cell, 23, 2131-2142. link
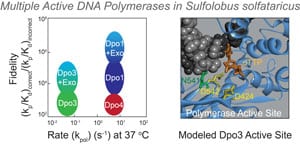
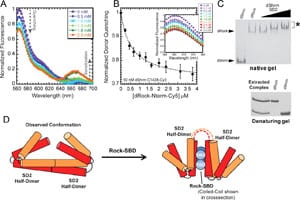 22) Bauer, R.J., Begley M.T., and Trakselis, M.A. (2012) Kinetics and fidelity of polymerization by DNA polymerase III from Sulfolobus solfataricus, Biochemistry, 51, 1996-2007. link
22) Bauer, R.J., Begley M.T., and Trakselis, M.A. (2012) Kinetics and fidelity of polymerization by DNA polymerase III from Sulfolobus solfataricus, Biochemistry, 51, 1996-2007. link
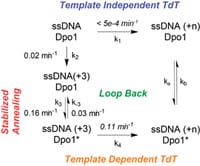
 21) Zuo, Z., Lin, H-K, and Trakselis, M.A. (2011) Strand annealing and terminal transferase activities of a B-family DNA Polymerase, Biochemistry, 50, 5379-5370. link
21) Zuo, Z., Lin, H-K, and Trakselis, M.A. (2011) Strand annealing and terminal transferase activities of a B-family DNA Polymerase, Biochemistry, 50, 5379-5370. link
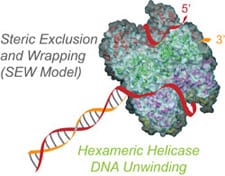
 20) Graham, B.W., Schauer, G.D., Leuba, S.H. and Trakselis, M.A. (2011) Steric Exclusion and Wrapping of the Excluded DNA Strand Occurs Along Discrete External Binding Paths During MCM Helicase Unwinding, NAR, 39, 6585. link
20) Graham, B.W., Schauer, G.D., Leuba, S.H. and Trakselis, M.A. (2011) Steric Exclusion and Wrapping of the Excluded DNA Strand Occurs Along Discrete External Binding Paths During MCM Helicase Unwinding, NAR, 39, 6585. link
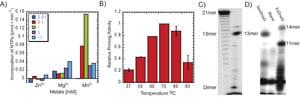
 19) Zuo, Z., Rodgers, C., Mikheikin, A.L., and Trakselis, M.A. (2010) Characterization of a functional DnaG-type primase in Archaea: Implications for a dual primase system, J. Mol. Biol., 397, 664-676. link
19) Zuo, Z., Rodgers, C., Mikheikin, A.L., and Trakselis, M.A. (2010) Characterization of a functional DnaG-type primase in Archaea: Implications for a dual primase system, J. Mol. Biol., 397, 664-676. link
 18) Mikheikin A.L., Lin, H-K, Mehta, P., Jen-Jacobson, L., and Trakselis, M.A. (2009) The DNA replication polymerase from Sulfolobus solfataricus forms a multimer when bound to DNA, NAR, 37, 7194-205. link
18) Mikheikin A.L., Lin, H-K, Mehta, P., Jen-Jacobson, L., and Trakselis, M.A. (2009) The DNA replication polymerase from Sulfolobus solfataricus forms a multimer when bound to DNA, NAR, 37, 7194-205. link
 17) Rothenberg, E., Trakselis, M.A., Bell, S.D., and Ha, T. (2007) MCM Forked substrate specificity involves dynamic interaction with the 5′-tail. JBC, 282, 34229-34. link
17) Rothenberg, E., Trakselis, M.A., Bell, S.D., and Ha, T. (2007) MCM Forked substrate specificity involves dynamic interaction with the 5′-tail. JBC, 282, 34229-34. link
From Gradute and Postdoc Work:
16) McGeoch, A.T., Trakselis, M.A., Laskey, R.A., and Bell, S.D. (2005) Organization of the Archaeal MCM Complex on DNA and Implications for a Helicase Mechanism. Nat Struc. Mol. Biol., 12, 756-762. (Cover) link
15) Trakselis, M.A., Alley, S.C., and Ishmael, F.T. (2005) Identification of Protein-Protein Interactions Using Novel Crosslinking Reagents. Bioconjugate Techniques, 16, 741-750. (Cover) link
14) Norcum, M.T., Warrington, A., Spiering, M.M., Ishmael, F.T., Trakselis, M.A., and Benkovic, S.J. (2005) Architecture of the Bacteriophage T4 Primosome: Electron Microscopy Studies of gp41 and gp61. PNAS, 102, 3623-3626. link
13) Zhang, Z, Spiering, M.M., Trakselis, M.A., Ishmael, F.T., Xi, J., Benkovic, S.J., and Hammes, G.G. (2005) Assembly of the Bacteriophage T4 Primosome: Single Molecule and Ensemble Studies. PNAS, 102, 3254-3259. link
12) Millar, D., Trakselis, M.A., and Benkovic, S.J. (2004) On the Solution of T4 Sliding Clamp (gp45). Biochemistry, 43, 12723-7. link
11) Zhang, Z., Spiering, M.M., Berdis, A.J., Trakselis, M.A., and Benkovic, S.J. (2004) ‘Screw-cap’ Clamp Loader Proteins that Thread. Nat. Struc. Biol. 11, 580-581. link
10) Trakselis, M.A. and Bell, S.D. (2004) Loader of the Rings. Nature, 249, 798-799. link
9) Yang, J., Zhuang, Z., Roccasecca, R., Trakselis, M.A., and Benkovic, S.J. (2004) The Dynamic Processivity of the T4 Polymerase during Replication. PNAS, 101, 8289-8294. link
8) Trakselis, M.A., Roccasecca, R., Yang, J., Valentine, A., and Benkovic, S.J. (2003) Dissociative Properties of the Proteins within the Bacteriophage T4 Replisome. JBC, 278, 49839-49849. link
7) Yang, J., Trakselis, M.A., Roccasecca, R., and Benkovic, S.J. (2003) The Application of a Minicircle Substrate in the Study of the Coordinated T4 DNA Replication. JBC, 278, 49828-49838. link
6) Ishmael, F.T. Trakselis, M.A., and Benkovic, S.J. (2003) Protein-Protein Interactions in the Bacteriophage T4 Replisome: The Leading Strand Holoenzyme is Physically Linked to the Lagging Strand Holoenzyme and Primosome. JBC, 278, 3145-3152. link
5) Trakselis, M.A., Berdis, A.J., Benkovic, S.J. (2003) Examination of the Role of the Clamp-Loader and ATP Hydrolysis in the Formation of the Bacteriophage T4 Polymerase Holoenzyme. J. Mol. Biol., 326, 435-451. link
4) Trakselis, M.A. and Benkovic, S.J. (2001) Intricacies in ATP Dependent Clamp Loading: Variations Across Replication Systems. Structure, 9, 999-1004. link
3) Alley, S.C., Trakselis, M.A., Mayer, M.U., Ishmael, F.T., Jones, A.D., Benkovic, S.J. (2001) Building a Replisome Solution Structure by Elucidation of Protein-Protein Interactions in the Bacteriophage T4 DNA Polymerase Holoenzyme. J. Biol. Chem., 276, 39340-39349. link
2) Trakselis, M.A., Mayer, M.U., Ishmael, F.T., Roccasecca R., and Benkovic, S.J. (2001) Use of Small Molecules to Determine Dynamic Protein-Protein Interactions in the Multi-Protein Bacteriophage T4 Replication Complex. TIBS, 26, 566-572. link
1) Trakselis, M.A., Alley, S.C., Abel-Santos, E., Benkovic, S.J. (2001) Creating a Dynamic Picture of the Sliding Clamp During T4 DNA Polymerase Holoenzyme Assembly Using Fluorescence Resonance Energy Transfer. PNAS, 98, 8368-8375. link