FDAAs
We developed a series of fluorescent probes, named fluorescent D-amino acids (FDAAs), for bacterial peptidoglycan labeling. These probes incorporate to peptidoglycan through the activity of cell wall synthases, indicating the formation and remodeling of peptidoglycan in situ.
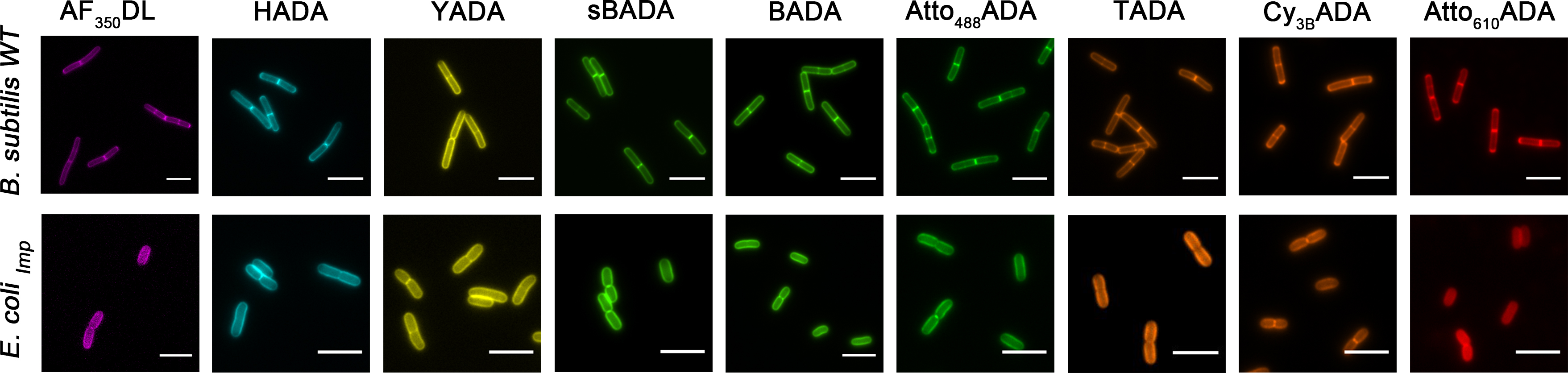
Figure 1 legends: FDAAs labeling in Bacillus subtilis (Gram-positive) and Escherichia coli (Gram-negative). Hsu et, al. 10.1039/C7SC01800B
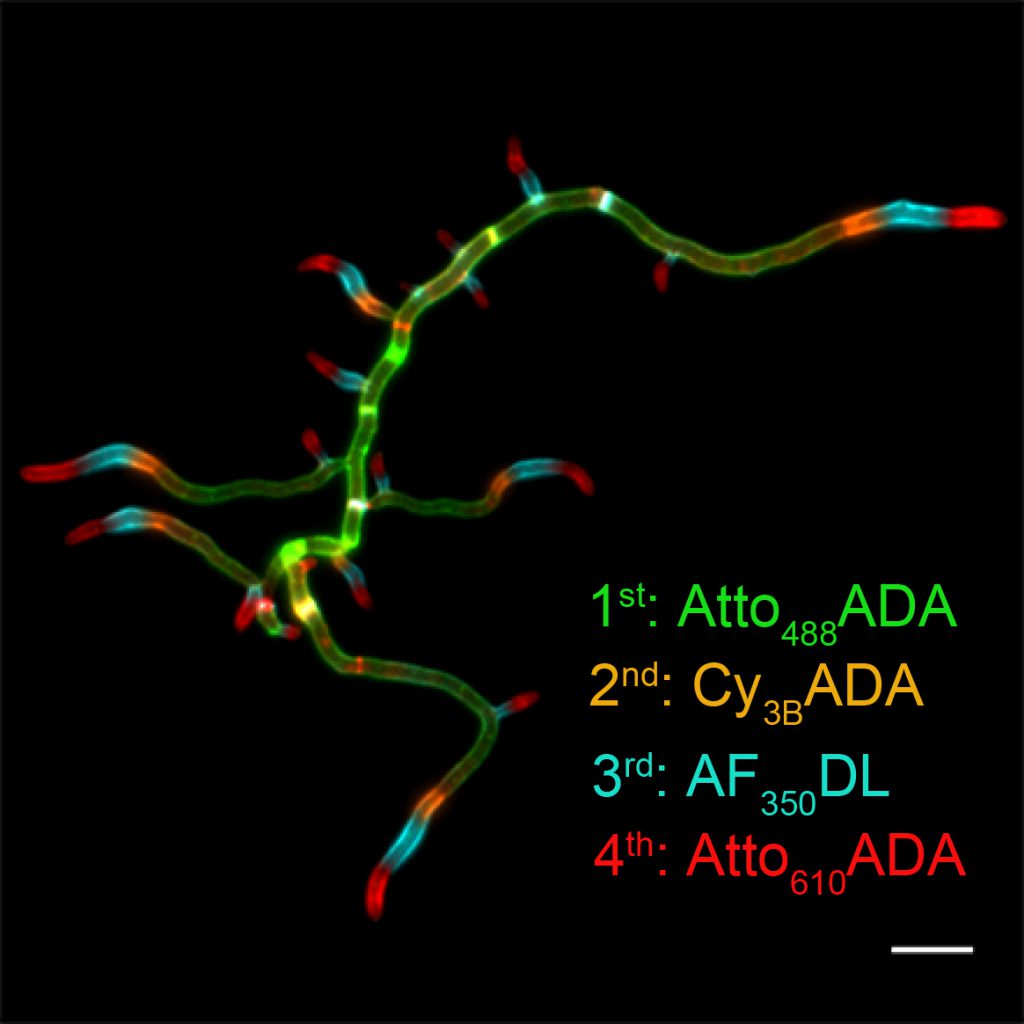
Figure 2 legends: Successive FDAA labeling in polar growing bacteria, Streptomyces venezuelae. This labeling method is also known as virtual time-lapse labeling. Hsu et, al. 10.1039/C7SC01800B
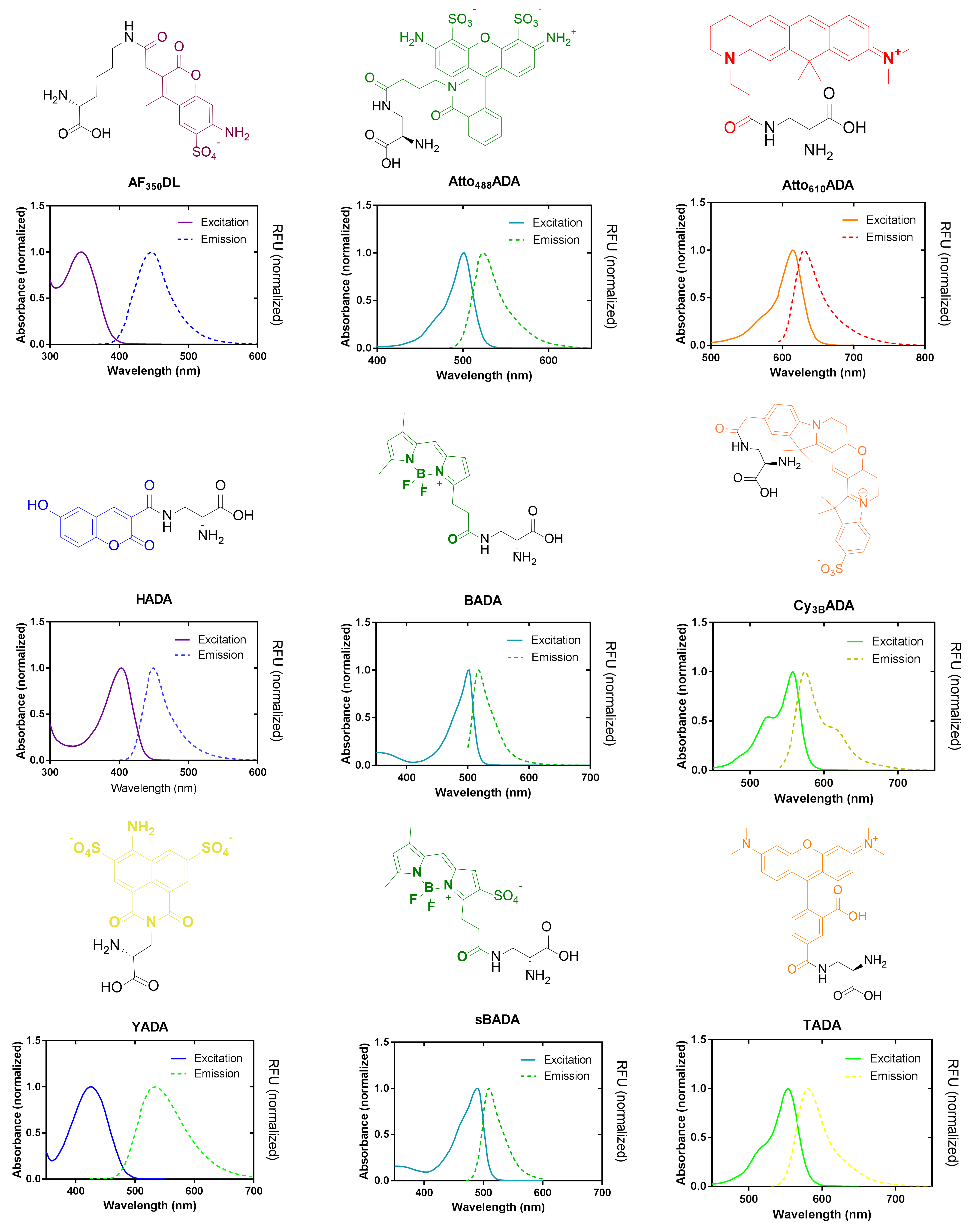
Figure 3 legends: Structures and spectra of FDAAs developed in the VanNieuwenhze Lab.
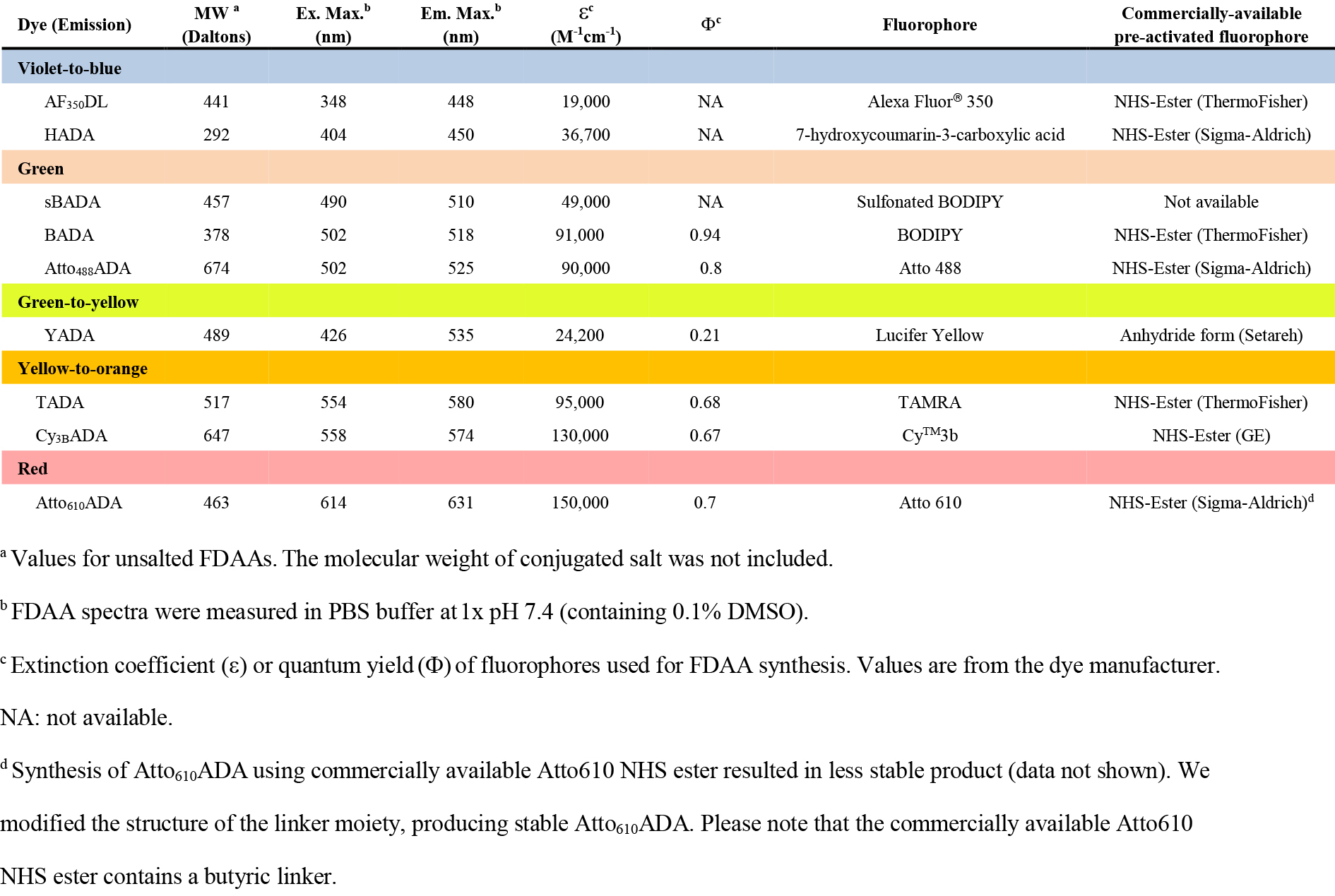
Figure 4 legends: Photochemical and physical properties of FDAAs. Hsu et, al. 10.1039/C7SC01800B
FDAA application and reference
First Author Research Title Journal/Year Developing D-amino acid-based probes for PG study E. Kuru In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids ACIEE 2012 M. Siegrist D-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen ACS Chem. Biol. 2012 M. Lebar Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis JACS 2014 E. Kuru Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ Nat. Protoc. 2014 J. Fura D-amino acid probes for penicillin binding protein-based bacterial surface labeling ASBMB 2015 S. Pidgeon Metabolic profiling of bacteria by unnatural c-terminated D-amino acids ACIEE 2015 S. Pidgeon Metabolic remodeling of bacterial surfaces via tetrazine ligations Chem. Commun. 2015 Y. Hsu Full color palette of fluorescent D‐amino acids for in situ labeling of bacterial cell walls Chem. Sci. 2017 Identifying the formation and/or structures of PG M. Pilhofer Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without Ftsz Nat. Commun. 2013 G. Billings De novo morphogenesis in L-forms via geometric control of cell growth Mol. Microbiol. 2014 G. Liechti A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis Nature 2014 M. van Teeseling Anammox planctomycetes have a peptidoglycan cell wall Nat. Commun. 2015 G. Liechti Pathogenic chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by mreb, for cell division PLoS Pathog. 2016 Studying bacterial morphogenesis and PG growth pattern E. Tocheva Peptidoglycan transformations during Bacillus subtilis sporulation Mol. Microbiol. 2013 A. Berezuk Site-directed fluorescence labeling reveals a revised n-terminal membrane topology and functional periplasmic residues in the escherichia coli cell division protein Ftsk J Biol. Chem. 2014 C. Kerr Salinity-dependent impacts of Proq, Prc, and Spr deficiencies on Escherichia coli cell structure J Bacteriol. 2014 A. Möll Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators J Bacteriol. 2014 T. Ursell Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization PNAS 2014 J. Monteiro Cell shape dynamics during the staphylococcal cell cycle Nat. Commun. 2015 H. Samal Molecular modeling, simulation and virtual screening of murd ligase protein from Salmonella typhimurium LT2 J Pharmacol. Toxicol. Methods 2015 M. Williams Short-stalked Prosthecomicrobium hirschii cells have a Caulobacter-like cell cycle J Bacteriol. 2016 A. Bisson-Filho Treadmilling by Ftsz filaments drives peptidoglycan synthesis and bacterial cell division Science 2017 E. Cserti Dynamics of the peptidoglycan biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium Mol. Microbiol. 2017 A. Dajkovic Hydrolysis of peptidoglycan is modulated by amidation of meso-diaminopimelic acid and Mg2+ in Bacillus subtilis Mol. Microbiol. 2017 H. Veiga Staphylococcus aureus requires at least one Ftsk/spoiiie protein for correct chromosome segregation Mol. Microbiol. 2017 X. Yang GTpase activity–coupled treadmilling of the bacterial tubulin Ftsz organizes septal cell wall synthesis Science 2017 Q. Yao Short Ftsz filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis EMBO J. 2017 Determining PG synthesis activity and sites Y. Eun Divin: a small molecule inhibitor of bacterial divisome assembly JACS 2013 T. Cameron Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division MBio. 2014 A. Fleurie Interplay of the serine/threonine-kinase Stkp and the paralogs DivIVA and Gpsb in Pneumococcal cell elongation and division PLoS Genet. 2014 A. Fleurie Mapz marks the division sites and positions Ftsz rings in Streptococcus pneumoniae Nature 2014 C. Jiang Sequential evolution of bacterial morphology by co-option of a developmental regulator Nature 2014 H. Tsui Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39 Mol. Microbiol. 2014 T. Dörr Endopeptidase-mediated beta lactam tolerance PLoS Pathog. 2015 B. Liu Roles for both Ftsa and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli Mol. Microbiol. 2015 A. Möll A D, D-carboxypeptidase is required for Vibrio cholerae halotolerance Environ. Microbiol. 2015 K. Sundararajan The bacterial tubulin Ftsz requires its intrinsically disordered linker to direct robust cell wall construction Nat. Commun. 2015 K. Schirner Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB Nat. Chem. Biol. 2015 A. Tavares MreC and MreD proteins are not required for growth of Staphylococcus aureus PLoS ONE 2015 A. Yakhnina The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability Mol. Microbiol. 2015 B. Jutras Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells PNAS 2016 S. Manuse Structure–function analysis of the extracellular domain of the Pneumococcal cell division site positioning protein MapZ Nat. Commun. 2016 A. Mura Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae J. Bacteriol. 2016 A. Pereira Ftsz-dependent elongation of a Coccoid bacterium MBio 2016 D. Ranjit Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid J Bacteriol. 2016 P. Yagȕe Subcompartmentalization by cross-membranes during early growth of Streptomyces hyphae Nat. Commun. 2016 D. Angeles Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site Mol. Microbiol. 2017 H. Botella Mycobacterium tuberculosis protease MarP activates a peptidoglycan hydrolase during acid stress EMBO J. 2017 L. Hamouche Bacillus subtilis swarmer cells lead the swarm, multiply, and generate a trail of quiescent descendants MBio 2017 A. Sugimoto Deciphering the mode of action of cell wall-inhibiting antibiotics using metabolic labeling of growing peptidoglycan in Streptococcus pyogenes Sci. Rep. 2017
Table 1. Examples of FDAAs applications in recent studies. Sub-headings classify the use of FDAAs in the experiment designs of each study.